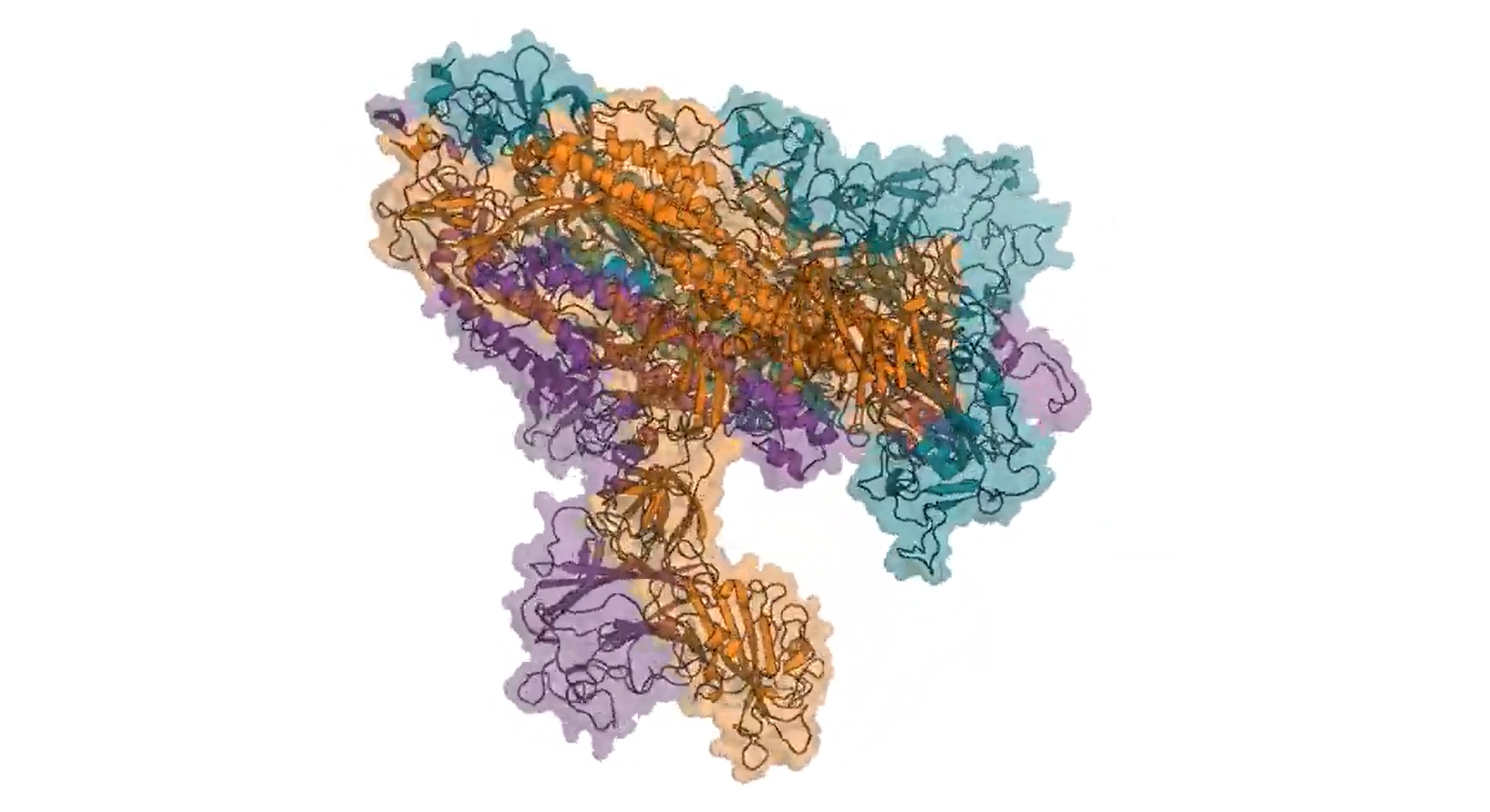
Molecular structure of the spike on the virus responsible for COVID-19, as simulated through Folding@home. The video from which this image is taken shows how the three proteins that make up the spike spread apart to reveal a binding site; this initiates infection by attaching to a particular protein on the surface of human cells.
CERN is contributing computing resources to a volunteer-computing initiative that aims to better understand the virus behind COVID-19. The initiative is being run on the Folding@home platform, which is a distributed computing project for simulating the dynamics of protein molecules. People across the globe can install software on their computers that will make use of spare processing power to help carry out complex simulations of how proteins fold and how their movements are implicated in a variety of diseases. The principle is the same as that which underpins CERN’s own volunteer-computing platform, LHC@home.
Like other viruses, the virus responsible for Covid-19 has proteins that are used to suppress hosts’ immune systems, as well as proteins for self-replication. Folding@home is supporting work to better understand these proteins, which is an important first step towards developing new drugs that could inhibit their function, thus stopping the virus.
While many methods exist for determining protein structures in the lab, these approaches reveal the shape at one given point in time. Computer simulations thus have an important role to play in determining how the three-dimensional structure of proteins can change over time. In the case of viral proteins, this can reveal new sites where drugs can be used to attack the protein and stop its functioning.
The number of people around the world contributing to Folding@home has grown rapidly over recent weeks. The maximum collective computing power of the system now stands at almost 2.5 exaFLOPS, which is more than that of the world’s top 500 supercomputers combined.
CERN is contributing about 10 000 computer cores from its main data centre. However, this only accounts for about a third of the ‘work units’ the CERN team has completed for Folding@home. The rest comes from contributions made directly by LHC computing sites. Taken all together, the CERN and LHC computing team has been the 87th biggest contributor as of 21 April.
“The contribution from the CERN data centre comes from machines that were due to be retired,” explains Jan van Eldik, leader of the Resource Provisioning section within the CERN IT department’s Compute and Monitoring group. “We have quickly developed a procedure to start virtual machines running Folding@home on CERN’s OpenStack cloud using these resources.”
“With the collaboration of the LHC experiments, we have shared similar ‘recipes’ — to maximise the contribution of available nodes to Folding@home — with computing sites across the Worldwide LHC Computing Grid (WLCG),” adds Simone Campana, WLCG project leader. This should make it easier for sites to contribute, in accordance with the agreements they have in place with their funding bodies.
“The Open Science Grid in the US is also a significant contributor to Folding@home,” notes Ian Bird, who preceded Campana as WLCG project leader from 2008 to January 2020. “Here at CERN, we have also offered to provide support for the server infrastructure of Folding@home, if needed.”
A number of research institutions are involved in running and enhancing Folding@home, which is led by Greg Bowman, an associate professor at Washington University School of Medicine in St. Louis in the US. Of course, Folding@home is just one of many large-scale distributed computing efforts that is working to help tackle COVID-19. Rosetta@home is another volunteer-computing initiative to which people can contribute from home. Important work is also being carried out in Europe: examples include work at data centres belonging to the Italian Institute for Nuclear Physics, and using the HADDOCK service on the WeNMR platform, which is supported by the EGI e-infrastructure.